This vignette describes how to use galamm to estimate
latent variable models with smooth terms, or equivalently, generalized
additive mixed models with factor structures. The examples are based on
Section 4 and 5 in Sørensen, Fjell, and Walhovd
(2023),
but as we cannot share the data, we have instead simulated somewhat
simpler datasets that will be used. We will gradually add complexity,
starting with a simple generalized additive mixed model. Please refer to
the introductory
vignette for an overview of the statistical models.
Generalized Additive Mixed Models
We start by showing how galamm can be used to estimated
generalized additive mixed models.
Gaussian Responses
The cognition dataset contains simulated data with
measurements of abilities in three cognitive domains.
head(cognition)
#> id domain x timepoint item trials y.y y.y.1
#> 1 1 1 0.06475113 1 11 1 0.16788973 1.00000000
#> 2 1 1 0.06475113 1 12 1 0.08897838 1.00000000
#> 3 1 1 0.06475113 1 13 1 0.03162123 1.00000000
#> 4 1 1 0.15766278 2 11 1 0.46598362 1.00000000
#> 5 1 1 0.15766278 2 12 1 0.84564656 1.00000000
#> 6 1 1 0.15766278 2 13 1 0.20549872 1.00000000For this first example, we focus only on the first item measured for the first domain.
dat <- subset(cognition, domain == 1 & item == "11")
dat$y <- dat$y[, 1]Each subject in this dataset has been measured eight times, and we can plot the measurements as follows:
plot(dat$x, dat$y, type = "n", xlab = "x", ylab = "y")
for(i in unique(dat$id)) {
dd <- dat[dat$id == i, ]
lines(dd$x, dd$y, col = "gray")
}
points(dat$x, dat$y, pch = 20, lwd = .05)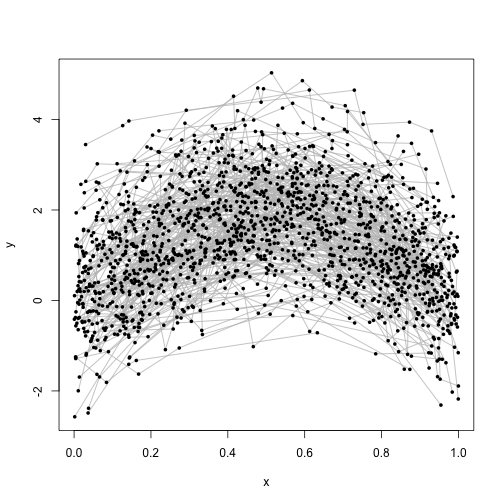
We use a generalized additive mixed model with random intercepts per subject to estimate the function relating to . In terms of the model framework outlined in the introductory vignette, we model the th response from the th subject with
where is a smooth function to be estimated, is a random intercept, and is a residual term.
This model can be estimated using gamm4 as follows:
The package gamm4 uses lme4 to fit the
underlying model, and the resulting model has two components.
mod_gamm4$mer contains the mixed model representation,
whereas in mod_gamm4$gam the fixed and random effects
corresponding to spline coefficients have been converted into single
smooth terms. We can look at the model summary for each:
summary(mod_gamm4$mer)
#> Linear mixed model fit by maximum likelihood ['lmerMod']
#>
#> AIC BIC logLik -2*log(L) df.resid
#> 3025.2 3052.1 -1507.6 3015.2 1595
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2.93755 -0.65215 0.00612 0.62654 3.14289
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 0.8551 0.9247
#> Xr s(x) 2.0341 1.4262
#> Residual 0.2501 0.5001
#> Number of obs: 1600, groups: id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> X(Intercept) 1.26938 0.06657 19.067
#> Xs(x)Fx1 -0.15814 0.20156 -0.785
#>
#> Correlation of Fixed Effects:
#> X(Int)
#> Xs(x)Fx1 0.000
summary(mod_gamm4$gam)
#>
#> Family: gaussian
#> Link function: identity
#>
#> Formula:
#> y ~ s(x)
#>
#> Parametric coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 1.26938 0.06657 19.07 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(x) 6.681 6.681 324.9 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.253
#> lmer.REML = 3015.2 Scale est. = 0.25012 n = 1600We can also plot the estimated smooth term:
plot(mod_gamm4$gam)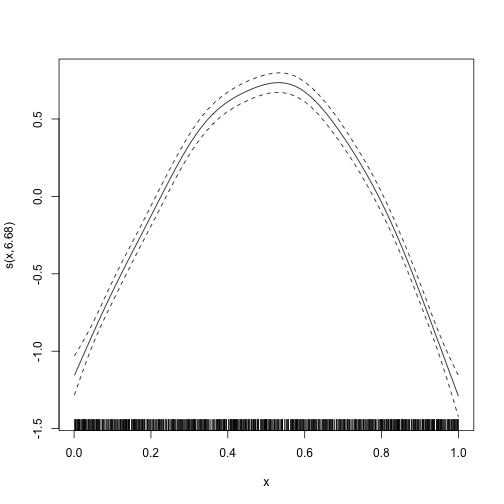
In contrast, invoking the plot function on the mixed
model part gives us a diagnostic plot.
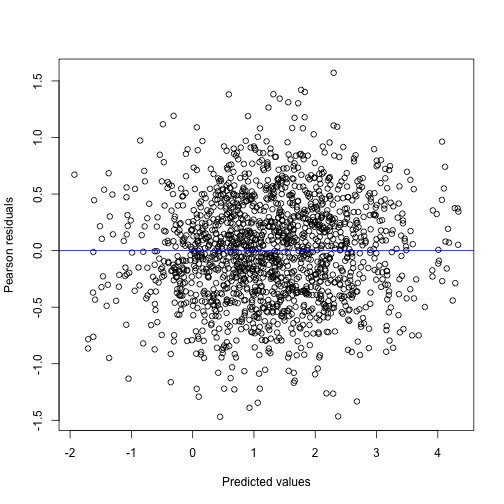
plot(mod_gamm4$mer)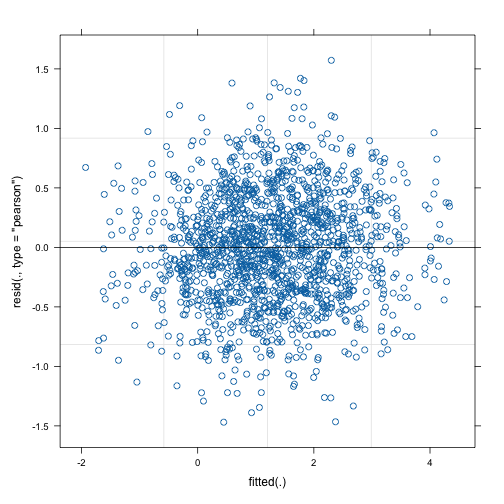
With galamm we use similar argument, but the
random specification is now part of the model formula.
As opposed to gamm4, galamm gives a single
summary. As can be seen, smooth terms are both reported as random
effects, and in a separate line under the header “Approximate
significance of smooth terms:”. Reassuringly, the results from fitting
the model with gamm4 and with galamm are
essentially equal, even though they use somewhat different computational
algorithms.
summary(mod)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ s(x) + (1 | id)
#> Data: dat
#>
#> AIC BIC logLik deviance df.resid
#> 3025.2 3052.1 -1507.6 3015.2 1595
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2.93755 -0.65215 0.00612 0.62654 3.14290
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 0.8551 0.9247
#> Xr s(x) 2.0346 1.4264
#> Residual 0.2501 0.5001
#> Number of obs: 1600, groups: id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 1.2694 0.06657 19.0672 4.731e-81
#> s(x)Fx1 -0.1582 0.20236 -0.7818 4.343e-01
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(x) 6.681 6.681 324.9 <2e-16The plot function now gives us a diagnostic plot, which
by inspection can be seen to be almost identical to the plot produced
from the mixed model part of the gamm4 model.
In order to plot the smooth term, we use
plot_smooth.
plot_smooth(mod)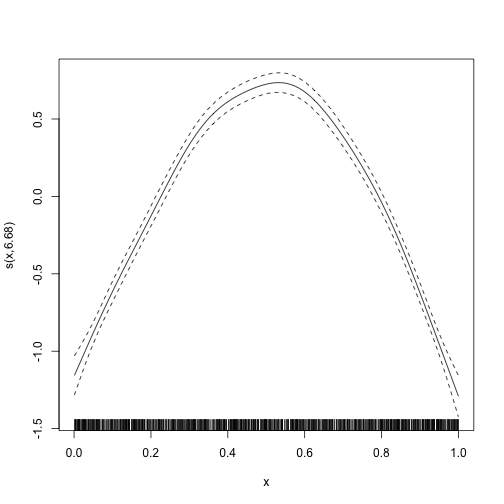
The plot_smooth function is a thin wrapper around the
plot.gam function provided by the mgcv package
(Wood 2017).
This means that the arguments used by plot.gam can be used
also here, as see with the examples below:
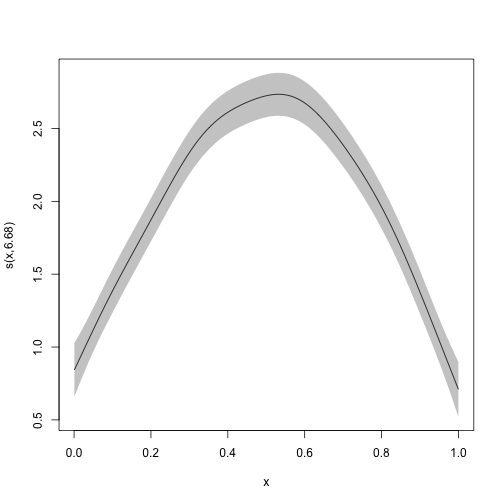
plot_smooth(mod,
shade = TRUE, rug = FALSE, seWithMean = TRUE,
shift = +2
)
plot_smooth(mod, se = FALSE)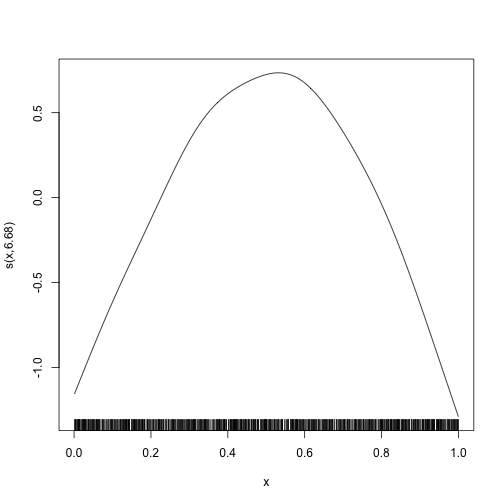
Binomial Responses
In the cognition dataset, the responses relating to domain 2 are binomially distributed. We will use the first trial to illustrate how such data can be modeled.
dat <- subset(cognition, domain == 2 & item == "21")
dat$y <- dat$y[, 1]Again we can fit this model using gamm4.
We can look at the summary output as before.
summary(mod_gamm4$mer)
#> Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
#> Family: binomial ( logit )
#>
#> AIC BIC logLik -2*log(L) df.resid
#> 983.7 1005.2 -487.8 975.7 1596
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -8.2548 0.0786 0.1946 0.4248 0.8213
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 0.2306 0.4802
#> Xr s(x) 0.9695 0.9846
#> Number of obs: 1600, groups: id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> X(Intercept) 2.8115 0.2034 13.825 < 2e-16 ***
#> Xs(x)Fx1 -1.4110 0.4242 -3.326 0.00088 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Correlation of Fixed Effects:
#> X(Int)
#> Xs(x)Fx1 -0.440
summary(mod_gamm4$gam)
#>
#> Family: binomial
#> Link function: logit
#>
#> Formula:
#> y ~ s(x)
#>
#> Parametric coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.8115 0.1662 16.91 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(x) 2.14 2.14 113.9 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.116
#> glmer.ML = 908.96 Scale est. = 1 n = 1600And we can plot the smooth term. The diagnostic plot is not very useful in the binomial case, so we omit it.
plot(mod_gamm4$gam)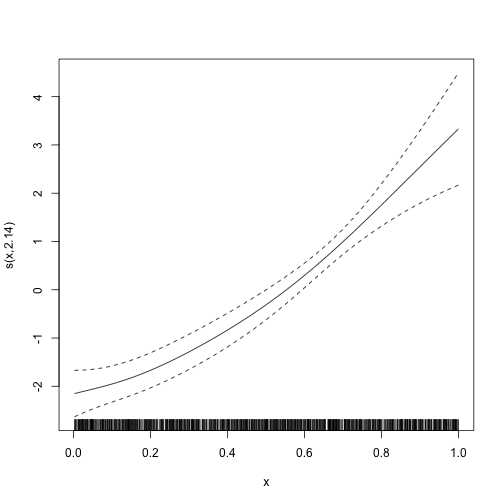
Again the galamm syntax is similar, but it puts the
random effect specification into the model formula.
The estimates are very similar, although not identical. The
difference in deviance is due to differences in the way deviance is
defined. The call deviance(mod_gamm4$mer) gives the same
value as in the summary for the model fitted with galamm.
summary(mod)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ s(x) + (1 | id)
#> Data: dat
#>
#> AIC BIC logLik deviance df.resid
#> 983.7 1005.2 -487.8 908.8 1596
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -8.2237 0.0792 0.1947 0.4245 0.8226
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 0.2316 0.4813
#> Xr s(x) 0.9387 0.9688
#> Number of obs: 1600, groups: id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.808 0.1956 14.354 9.989e-47
#> s(x)Fx1 -1.406 0.4134 -3.402 6.697e-04
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(x) 2.124 2.124 114 <2e-16
plot_smooth(mod)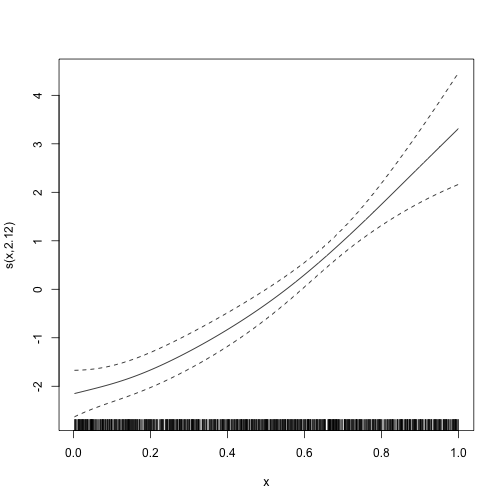
Generalized Additive Models with Factor Structures
We now add factor structures to the GAMMs. These are the types of
models that neither gamm4 nor mgcv are able to
estimate (at least without lots of manual hacking), and where
galamm provides new functionality.
Gaussian Responses
To illustrate basic usage, we continue with the cognition data, but now use all items of cognitive domain 1. These are all conditionally normal distributed.
dat <- subset(cognition, domain == 1)
dat$y <- dat$y[, 1]
head(dat)
#> id domain x timepoint item trials y
#> 1 1 1 0.06475113 1 11 1 0.16788973
#> 2 1 1 0.06475113 1 12 1 0.08897838
#> 3 1 1 0.06475113 1 13 1 0.03162123
#> 4 1 1 0.15766278 2 11 1 0.46598362
#> 5 1 1 0.15766278 2 12 1 0.84564656
#> 6 1 1 0.15766278 2 13 1 0.20549872We now need a factor model to associate the underlying latent trait with the measurements :
In the structural model, we have a smooth term for the relationship between the latent trait and x, and we have random intercepts for a given timepoint within subject , and for a given subject across timepoints .
The reduced form of the model is
We will use a varying-coefficient term, where is being interpreted as a regression coefficient for the effect of on , and the regression term varies with . In contrast to Hastie and Tibshirani (1993) and other uses of varying-coefficient terms, however, in this case the predictor is a model parameter. We have three items loading in and fix the first loading to 1 for identifiability, so the loading matrix is as follows:
We provide thin wrappers around the s() and
t2() functions from mgcv to support factor
loadings in smooth terms. The wrappers are named sl() and
t2l() to avoid namespace conflicts with mgcv
and gamm4, and the last letter “l” stands for “loading”. In
this example, we set factor = "item" to specify that the
loadings to be applied are identified by the “item” variable. Using
mgcv’s by variable would also work in this
particular case, i.e., replacing sl(x, factor = "loading")
with s(x, by = loading). However, in most cases this would
lead to identifiability issues due to the way varying-coefficient terms
are set up by mgcv, so galamm provides an
additional factor argument which alleviates most of these
issues.
mod <- galamm(
formula = y ~ 0 + item + sl(x, factor = "loading") +
(0 + loading | id / timepoint),
data = dat,
load_var = "item",
lambda = loading_matrix,
factor = "loading"
)We print the model summary below. In the data simulation, the factor
loadings were set to 1, 1.4, and 0.3, respectively, and this is very
well recovered. Furthermore, the ground truth standard deviation at the
id level was 1, at the timepoint level it was
0.5, and the residual standard deviation was 0.1. The estimates are
close to these values. Real data will typically not have this strong
signal, but based on these results, there are no clear indications that
the model is implemented incorrectly.
summary(mod)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ 0 + item + sl(x, factor = "loading") + (0 + loading | id/timepoint)
#> Data: dat
#>
#> AIC BIC logLik deviance df.resid
#> -918.2 -853.4 469.1 -938.2 4790
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -92.78 -65.29 -50.78 -17.21 -10.48
#>
#> Lambda:
#> loading SE
#> lambda1 1.0000 .
#> lambda2 1.3973 0.003531
#> lambda3 0.3009 0.002146
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> timepoint:id loading 0.236886 0.48671
#> id loading 0.857051 0.92577
#> Xr s(x):loading 2.030613 1.42500
#> Residual 0.009932 0.09966
#> Number of obs: 4800, groups: timepoint:id, 1600; id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error t value Pr(>|t|)
#> item11 1.2694 0.06663 19.0513 6.412e-81
#> item12 1.7788 0.09307 19.1128 1.977e-81
#> item13 0.3797 0.02019 18.8077 6.531e-79
#> s(x):loadingFx1 0.1496 0.19977 0.7488 4.540e-01
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(x):loading 8.719 8.719 4469 <2e-16We also plot the smooth term. Since we had a very large amount of data, there is essentially no uncertainty about the estimate.
plot_smooth(mod)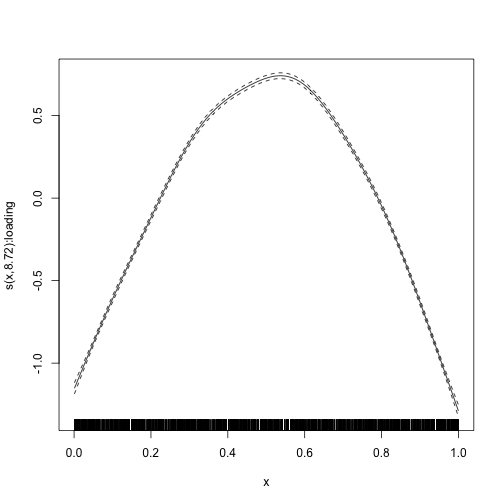
Binomial Responses
We can now move on to the part of the cognition data that is
conditionally binomially distributed. We consider domain 2, where each
response measures success or not in a single trial. In this case there
are only two items, so we must change the lambda matrix accordingly.
Other than that, and setting family = binomial, the model
is the same as before.
dat <- subset(cognition, domain == 2)
dat$y <- dat$y[, 1]
mod <- galamm(
formula = y ~ 0 + item + sl(x, factor = "loading") +
(0 + loading | id / timepoint),
data = dat,
family = binomial,
load_var = "item",
lambda = matrix(c(1, NA), ncol = 1),
factor = "loading"
)The summary is shown below. The factor loading
was used when simulating the data, and including the uncertainty, our
estimate covers the true value well. Also note that the variation
between individuals (group id) and the variation between
timepoints within individuals (group timepoint:id) gets
lumped together at the id level. The estimated variation at
the timepoint:id level is zero. This is a well-known
phenomenon when fitting mixed models, given book-length treatment in
Hodges (2013). In
this case, it is likely due to the fact that we only have two
measurements at each timepoint, and also the fact that we use the
Laplace approximation to integrate over the random effects, and this
approximation may be inaccurate for binomial data with a low number of
repeated observations (Joe
2008).
summary(mod)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ 0 + item + sl(x, factor = "loading") + (0 + loading | id/timepoint)
#> Data: dat
#>
#> AIC BIC logLik deviance df.resid
#> 1495.5 1538.0 -740.8 1614.8 3193
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -15.8546 0.0279 0.0780 0.1985 1.3948
#>
#> Lambda:
#> loading SE
#> lambda1 1.000 .
#> lambda2 2.202 0.3007
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> timepoint:id loading 0.0000 0.0000
#> id loading 0.6222 0.7888
#> Xr s(x):loading 1.5388 1.2405
#> Number of obs: 3200, groups: timepoint:id, 1600; id, 200; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> item21 2.944 0.1903 15.473 5.249e-54
#> item22 6.319 0.5853 10.796 3.612e-27
#> s(x):loadingFx1 -1.389 0.2837 -4.897 9.733e-07
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(x):loading 2.491 2.491 115.1 <2e-16The true value 2 for the factor loading is well within the 95 % confidence limits.
confint(mod, parm = "lambda")
#> 2.5 % 97.5 %
#> lambda1 1.612341 2.791192Smooth Terms with Loadings and Factor Interactions
Gaussian Responses
Domain 1 and 3 both have Gaussian responses, and we can model them jointly.
We also add indicator variables for the two domains.
dat <- cbind(
dat,
model.matrix(~ 0 + domain, data = dat)[, c("domain1", "domain3")]
)We define the loading matrix, now having two columns:
(lmat <- matrix(c(
1, NA, NA, 0, 0, 0, 0,
0, 0, 0, 1, NA, NA, NA
), ncol = 2))
#> [,1] [,2]
#> [1,] 1 0
#> [2,] NA 0
#> [3,] NA 0
#> [4,] 0 1
#> [5,] 0 NA
#> [6,] 0 NA
#> [7,] 0 NAThen we define the model. The smooth term is now
sl(x, by = domain, factor = c("ability1", "ability3")),
indicating that there should be a separate smooth term for each level of
domain, and that the term should be multiplied by the
loading “ability1” or “ability3”. We also set factr = 1e9
to be less strict with regards to convergence than usual, because this
model is hard to estimate.
mod_byvar <- galamm(
formula = y ~ domain +
sl(x, by = domain, factor = c("ability1", "ability3")) +
(0 + domain1:ability1 + domain3:ability3 | id / timepoint),
data = dat,
load_var = "item",
lambda = lmat,
factor = c("ability1", "ability3"),
control = galamm_control(
optim_control = list(factr = 1e9, trace = 3,
REPORT = 50, maxit = 1000)
)
)
#> N = 17, M = 20 machine precision = 2.22045e-16
#> At X0, 0 variables are exactly at the bounds
#> At iterate 0 f= 25324 |proj g|= 9642.6
#> At iterate 50 f = 136.9 |proj g|= 716.66
#> At iterate 100 f = -1036.1 |proj g|= 2364.3
#> At iterate 150 f = -1069 |proj g|= 46.6
#> At iterate 200 f = -1147.4 |proj g|= 95.18
#> At iterate 250 f = -1217.6 |proj g|= 53.837
#>
#> iterations 266
#> function evaluations 292
#> segments explored during Cauchy searches 269
#> BFGS updates skipped 0
#> active bounds at final generalized Cauchy point 0
#> norm of the final projected gradient 8.60877
#> final function value -1217.8
#>
#> F = -1217.8
#> final value -1217.796853
#> convergedThe summary shows that we have recovered the true values of the factor loadings well.
summary(mod_byvar)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ domain + sl(x, by = domain, factor = c("ability1", "ability3")) +
#> (0 + domain1:ability1 + domain3:ability3 | id/timepoint)
#> Data: dat
#> Control: galamm_control(optim_control = list(factr = 1e+09, trace = 3, REPORT = 50, maxit = 1000))
#>
#> AIC BIC logLik deviance df.resid
#> -2399.6 -2267.8 1217.8 -2435.6 11182
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2115.29 -1034.70 -885.48 21.85 50.66
#>
#> Lambda:
#> ability1 SE ability3 SE
#> lambda1 1.0000 . . .
#> lambda2 1.3986 0.002569 . .
#> lambda3 0.3013 0.002007 . .
#> lambda4 . . 1.0000 .
#> lambda5 . . 0.9993 0.0007676
#> lambda6 . . 1.0001 0.0007679
#> lambda7 . . 1.9995 0.0015686
#>
#> Random effects:
#> Groups Name Variance Std.Dev. Corr
#> timepoint:id domain1:ability1 0.23651 0.4863
#> domain3:ability3 0.28043 0.5296 -0.01
#> id domain1:ability1 3.35714 1.8322
#> domain3:ability3 18.34082 4.2826 0.88
#> Xr.0 s(x):domain3:ability3 87.28287 9.3425
#> Xr s(x):domain1:ability1 14.27050 3.7776
#> Residual 0.01007 0.1003
#> Number of obs: 11200, groups: timepoint:id, 1600; id, 200; Xr.0, 8; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -0.004819 0.004671 -1.0318 3.022e-01
#> domain3 0.002595 0.007592 0.3418 7.325e-01
#> s(x):domain1:ability1Fx1 -0.065816 0.241553 -0.2725 7.853e-01
#> s(x):domain3:ability3Fx1 5.569172 0.314765 17.6931 4.739e-70
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(x):domain1:ability1 8.951 8.951 4316 <2e-16
#> s(x):domain3:ability3 8.988 8.988 59836 <2e-16We can plot the estimated smooth terms, which recover their simulated ground truth very well.
plot_smooth(mod_byvar, scale = 0, select = 1)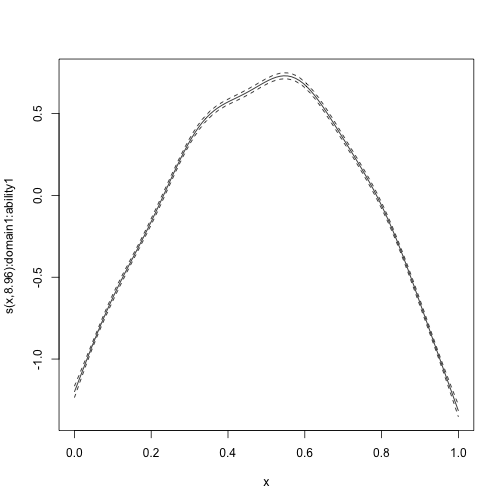
plot_smooth(mod_byvar, scale = 0, select = 2)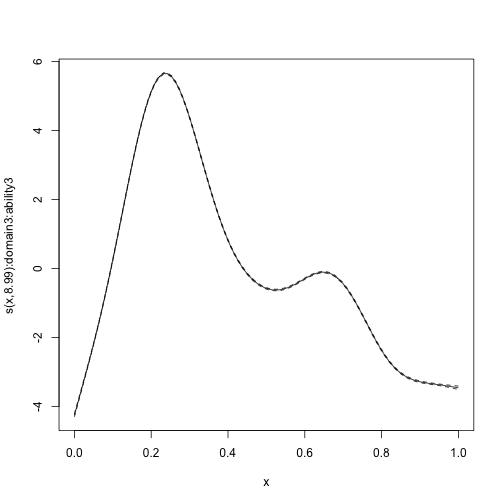
Mixed Gaussian and Binomial Responses
Domain 1 has Gaussian responses and domain 2 has binomial responses, and we can model them jointly. For the sake of speed, we include only two items for each domain.
We also add indicator variables for the two domains.
dat <- cbind(
dat,
model.matrix(~ 0 + domain, data = dat)[, c("domain1", "domain2")]
)We define the loading matrix, now having two columns:
(lmat <- matrix(c(
1, NA, NA, 0, 0,
0, 0, 0, 1, NA
), ncol = 2))
#> [,1] [,2]
#> [1,] 1 0
#> [2,] NA 0
#> [3,] NA 0
#> [4,] 0 1
#> [5,] 0 NAThen we define the model. The smooth term is now
sl(x, by = domain, factor = c("ability1", "ability2")),
indicating that there should be a separate smooth term for each level of
domain, and that the term should be multiplied by the
loading “ability1” or “ability2”. Because this model has some
convergence issues, we omit the timepoint-level random intercepts in
this example. Since gfam is also exported from
gamm4, which we loaded at the top of this vignette, we use
galamm::gfam to get the correct function.
mod_byvar_mixed <- galamm(
formula = y ~ domain +
sl(x, by = domain, factor = c("ability1", "ability2")) +
(0 + domain1:ability1 + domain2:ability2 | id),
data = dat,
family = galamm::gfam(list(gaussian, binomial)),
load_var = "item",
lambda = lmat,
factor = c("ability1", "ability2"),
control = galamm_control(
optim_control = list(factr = 1e9, trace = 3,
REPORT = 30, maxit = 1000)
)
)
#> N = 12, M = 20 machine precision = 2.22045e-16
#> At X0, 0 variables are exactly at the bounds
#> At iterate 0 f= 8170.1 |proj g|= 2305.5
#> At iterate 30 f = 4726.2 |proj g|= 39.814
#>
#> iterations 44
#> function evaluations 49
#> segments explored during Cauchy searches 44
#> BFGS updates skipped 0
#> active bounds at final generalized Cauchy point 0
#> norm of the final projected gradient 0.286444
#> final function value 4725.04
#>
#> F = 4725.04
#> final value 4725.035302
#> convergedWe can look at the model summary:
summary(mod_byvar_mixed)
#> GALAMM fit by maximum marginal likelihood.
#> Formula: y ~ domain + sl(x, by = domain, factor = c("ability1", "ability2")) +
#> (0 + domain1:ability1 + domain2:ability2 | id)
#> Data: dat
#> Control: galamm_control(optim_control = list(factr = 1e+09, trace = 3, REPORT = 30, maxit = 1000))
#>
#> AIC BIC logLik deviance df.resid
#> 9476.1 9566.9 -4725.0 58315.3 7987
#>
#> Lambda:
#> ability1 SE ability2 SE
#> lambda1 1.0000 . . .
#> lambda2 1.4082 0.01335 . .
#> lambda3 0.2869 0.01040 . .
#> lambda4 . . 1.0000 .
#> lambda5 . . 0.7996 0.05898
#>
#> Random effects:
#> Groups Name Variance Std.Dev. Corr
#> id domain1:ability1 2.4081 1.552
#> domain2:ability2 1.4827 1.218 0.40
#> Xr.0 s(x):domain2:ability2 0.8501 0.922
#> Xr s(x):domain1:ability1 2.2392 1.496
#> Number of obs: 8000, groups: id, 200; Xr.0, 8; Xr, 8
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.04306 0.02253 1.9116 5.593e-02
#> domain2 3.23013 0.16886 19.1290 1.449e-81
#> s(x):domain1:ability1Fx1 -0.08920 0.13855 -0.6438 5.197e-01
#> s(x):domain2:ability2Fx1 2.10045 0.47649 4.4082 1.042e-05
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(x):domain1:ability1 7.865 7.865 880 <2e-16
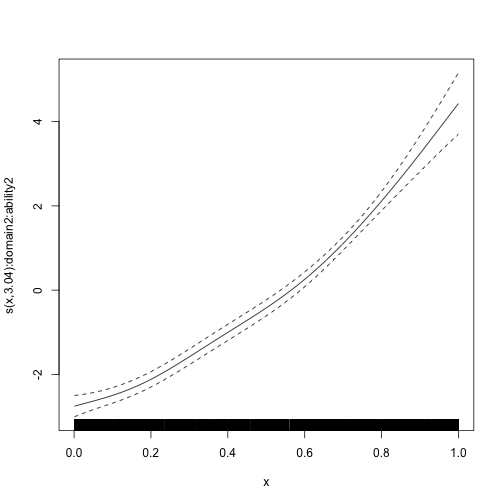
#> s(x):domain2:ability2 3.036 3.036 285 <2e-16We can plot the estimated smooth terms:
plot_smooth(mod_byvar_mixed, scale = 0, select = 1)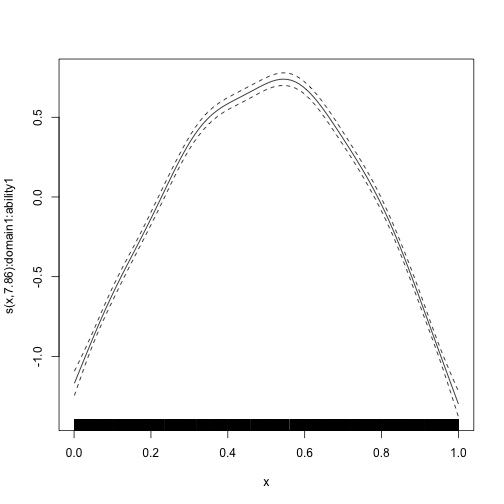
plot_smooth(mod_byvar_mixed, scale = 0, select = 2)